Work with STAC catalogs on the SURF dCache Storage¶
Create a STAC catalog¶
We search for some assets in the Sentinel-2 Open Data collection available on AWS. We query the Earth Search STAC API end point using PySTAC Client. In order to install this tool, uncomment and run the following cell:
[1]:
# !pip install pystac-client
[2]:
from pystac_client import Client
[3]:
STAC_API_URL = "https://earth-search.aws.element84.com/v0"
client = Client.open(STAC_API_URL)
# search assets
search = client.search(
collections=["sentinel-s2-l2a-cogs"],
datetime="2018-03-16/2018-03-25",
# query Sentinel-2 tile 5VNK
query=[
"sentinel:utm_zone=5",
"sentinel:latitude_band=V",
"sentinel:grid_square=NK"
]
)
[4]:
# get all items matching the query
items = search.get_all_items()
We then use PySTAC to create a STAC catalog where to save the search results.
[5]:
from pystac import Catalog, Item
[6]:
# create new catalog
catalog = Catalog(
id='s2-catalog',
description='Test catalog for Sentinel-2 data'
)
catalog
[6]:
Catalog: s2-catalog
| ID: s2-catalog |
| Description: Test catalog for Sentinel-2 data |
Links
Link:
| Rel: root |
| Target: |
| Media Type: application/json |
[7]:
# add search results to catalog
catalog.add_items(items)
catalog.describe()
* <Catalog id=s2-catalog>
* <Item id=S2B_5VNK_20180325_1_L2A>
* <Item id=S2A_5VNK_20180324_0_L2A>
* <Item id=S2B_5VNK_20180322_0_L2A>
* <Item id=S2B_5VNK_20180319_0_L2A>
Read/write STAC objects¶
Let’s save the catalog (metadata only!) on the dCache storage. For authentication we use a macaroon (see here for instructions on how to generate the token), but username/password authentication can also be employed:
[8]:
import stac2dcache
stac2dcache.configure(token_filename="macaroon.dat")
We can then read/write PySTAC objects on dCache as:
[9]:
urlpath = "https://webdav.grid.surfsara.nl:2880/pnfs/grid.sara.nl/data/eratosthenes/disk/tmp-sentinel-2-catalog"
# temporary fix for pystac<1.4
catalog._stac_io = stac2dcache.stac_io
catalog.normalize_and_save(
urlpath,
catalog_type='SELF_CONTAINED'
)
The catalog is now written to the storage. In order to re-load it into memory:
[10]:
catalog_url = urlpath + '/catalog.json'
# read catalog from storage
catalog = Catalog.from_file(
catalog_url,
stac_io=stac2dcache.stac_io
)
[11]:
catalog.describe()
* <Catalog id=s2-catalog>
* <Item id=S2B_5VNK_20180325_1_L2A>
* <Item id=S2A_5VNK_20180324_0_L2A>
* <Item id=S2B_5VNK_20180322_0_L2A>
* <Item id=S2B_5VNK_20180319_0_L2A>
Retrieve assets to/from dCache¶
Let’s now retrieve few assets from the remote servers, and save them to dCache. We download the original XML metadata file and one band file (‘B-1’) from the MSI.
[12]:
from stac2dcache.utils import copy_asset
[13]:
# download assets - from web to storage
for asset_key in ('metadata', 'B01'):
copy_asset(
catalog,
asset_key,
update_catalog=True, # update the catalog's links to the assets
filesystem_to=stac2dcache.fs,
max_workers=4
)
# save catalog with the updated links
catalog.normalize_and_save(urlpath)
Note that copy_asset makes use of multiple processes to download the data (use the max_workers argument to set the number of processes spawned).
Files have been saved on the dCache storage. Let’s now download one example file from dCache to the local file system for inspection or further processing.
[14]:
# download an asset - from storage to local filesystem
copy_asset(
catalog,
'B01',
item_id='S2B_5VNK_20180319_0_L2A',
to_uri='./tmp',
filesystem_from=stac2dcache.fs
)
[15]:
! ls ./tmp/S2B_5VNK_20180319_0_L2A/B01.tif
./tmp/S2B_5VNK_20180319_0_L2A/B01.tif
Load assets¶
Instead of copying assets locally, STAC2dCache allows to directly load into memory few common file formats:
[16]:
from stac2dcache.utils import get_asset
[17]:
# read metadata from storage (as text file)
data = get_asset(
catalog,
'metadata',
'S2B_5VNK_20180319_0_L2A',
filesystem=stac2dcache.fs
)
# print top five lines
data.splitlines()[:5]
[17]:
["<?xml version='1.0' encoding='UTF-8'?>",
'<n1:Level-2A_Tile_ID xmlns:n1="https://psd-14.sentinel2.eo.esa.int/PSD/S2_PDI_Level-2A_Tile_Metadata.xsd" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" xsi:schemaLocation="https://psd-14.sentinel2.eo.esa.int/PSD/S2_PDI_Level-2A_Tile_Metadata.xsd /dpc/app/s2ipf/FORMAT_METADATA_TILE_L1C/02.11.01/scripts/../../../schemas/02.13.01/PSD/S2_PDI_Level-2A_Tile_Metadata.xsd">',
' <n1:General_Info>',
' <L1C_TILE_ID metadataLevel="Brief">S2B_OPER_MSI_L1C_TL_EPAE_20180319T231157_A005403_T05VNK_N02.06</L1C_TILE_ID>',
' <TILE_ID metadataLevel="Brief">S2B_OPER_MSI_L2A_TL_SHIT_20201104T123444_A005403_T05VNK_N00.01</TILE_ID>']
[18]:
# same for raster data (as xarray object)
data = get_asset(
catalog,
'B01',
'S2B_5VNK_20180319_0_L2A',
filesystem=stac2dcache.fs
)
data
[18]:
<xarray.DataArray (band: 1, y: 1830, x: 1830)>
array([[[ 0, 0, 0, ..., 5703, 5936, 5016],
[ 0, 0, 0, ..., 6587, 6828, 5825],
[ 0, 0, 0, ..., 6699, 6655, 5496],
...,
[ 9110, 10489, 9932, ..., 1733, 2269, 1933],
[ 9406, 10773, 10684, ..., 1799, 2759, 2406],
[ 8260, 9473, 9653, ..., 1170, 1634, 1917]]], dtype=uint16)
Coordinates:
* band (band) int64 1
* x (x) float64 5e+05 5.001e+05 5.001e+05 ... 6.097e+05 6.098e+05
* y (y) float64 7e+06 7e+06 7e+06 ... 6.89e+06 6.89e+06 6.89e+06
spatial_ref int64 0
Attributes:
AREA_OR_POINT: Area
OVR_RESAMPLING_ALG: AVERAGE
_FillValue: 0
scale_factor: 1.0
add_offset: 0.0In order to plot the data, matplotlib needs to be installed (uncomment and run the following lines to do that).
[19]:
# !pip install matplotlib
[20]:
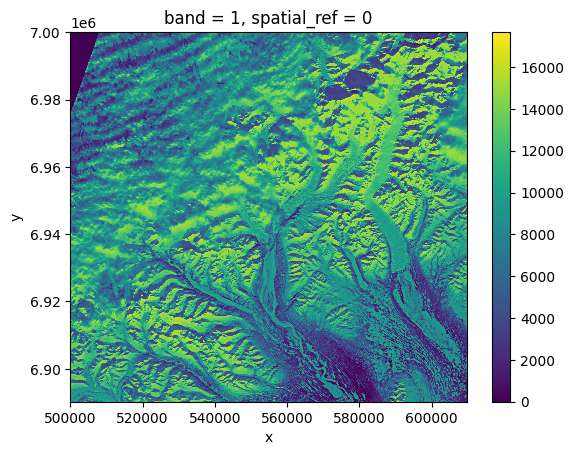
data.plot()
[20]:
<matplotlib.collections.QuadMesh at 0x7f8c00e5d730>
Use a configuration file¶
The token for authentication, together with other parameters (such as the dCache WebDAV door), can be saved in a .json (or .ini) configuration file stored in the directory ~/.config/fsspec/:
{
"dcache": {
"api_url": "https://dcacheview.grid.surfsara.nl:22880/api/v1",
"webdav_url": "https://webdav.grid.surfsara.nl:2880",
"token": "<DCACHE_TOKEN>",
"block_size": 0
}
}
Once setup as above, STAC2dCache will be able to read/write URL-paths that include the “dcache” protocol without specifying the stac_io/filesystem arguments:
[21]:
catalog_url = "dcache://pnfs/grid.sara.nl/data/eratosthenes/disk/tmp-sentinel-2-catalog/catalog.json"
# read catalog from storage
catalog = Catalog.from_file(catalog_url)
catalog.describe()
* <Catalog id=s2-catalog>
* <Item id=S2B_5VNK_20180325_1_L2A>
* <Item id=S2A_5VNK_20180324_0_L2A>
* <Item id=S2B_5VNK_20180322_0_L2A>
* <Item id=S2B_5VNK_20180319_0_L2A>
[22]:
data = get_asset(
catalog,
'B01',
'S2B_5VNK_20180319_0_L2A'
)
data
[22]:
<xarray.DataArray (band: 1, y: 1830, x: 1830)>
array([[[ 0, 0, 0, ..., 5703, 5936, 5016],
[ 0, 0, 0, ..., 6587, 6828, 5825],
[ 0, 0, 0, ..., 6699, 6655, 5496],
...,
[ 9110, 10489, 9932, ..., 1733, 2269, 1933],
[ 9406, 10773, 10684, ..., 1799, 2759, 2406],
[ 8260, 9473, 9653, ..., 1170, 1634, 1917]]], dtype=uint16)
Coordinates:
* band (band) int64 1
* x (x) float64 5e+05 5.001e+05 5.001e+05 ... 6.097e+05 6.098e+05
* y (y) float64 7e+06 7e+06 7e+06 ... 6.89e+06 6.89e+06 6.89e+06
spatial_ref int64 0
Attributes:
AREA_OR_POINT: Area
OVR_RESAMPLING_ALG: AVERAGE
_FillValue: 0
scale_factor: 1.0
add_offset: 0.0More info on the use of configuration files for the underlying Filesystem Spec library here.
Retrieve assets from API endpoints that require authentication¶
Below we demontrate how to search Sentinel-1 GRD data from the Alaska Satellite Facility (ASF) enpoint that is part of the NASA Common Metadata Repository (CMR) STAC catalog. While the catalog is publicly accessible, authentication is required in order to download the assets.
As for Sentinel-2, we first search the available assets with PySTAC Client:
[23]:
STAC_API_URL = "https://cmr.earthdata.nasa.gov/stac/ASF"
bbox = [4.6, 52.3, 4.7, 52.4] # (min lon, min lat, max lon, max lat) AMS area
client = Client.open(STAC_API_URL)
# search assets
search = client.search(
collections=[
"SENTINEL-1A_DP_GRD_HIGH.v1",
"SENTINEL-1B_DP_GRD_HIGH.v1"
],
datetime="2019-01-02",
bbox=bbox
)
[24]:
# get all items matching the query
items = search.get_all_items()
[25]:
len(items)
[25]:
1
We build a catalog with the found items. Note that we have limited our search to a single item due to the large size of S1 GRD data.
[26]:
catalog = Catalog(
id='s1-catalog',
description='Test catalog for Sentinel-1 data'
)
catalog.add_items(items)
catalog.describe()
* <Catalog id=s1-catalog>
* <Item id=S1B_IW_GRDH_1SDV_20190102T055723_20190102T055748_014311_01AA09_29B6-GRD_HD>
[27]:
urlpath = "dcache://pnfs/grid.sara.nl/data/eratosthenes/disk/tmp-sentinel-1-catalog"
catalog.normalize_and_save(
urlpath,
catalog_type='SELF_CONTAINED'
)
To retrieve Sentinel-1 images, we need to authenticate using EarthData login credentials (see this guide for instructions on how to register). Also make sure you authorize all applications related to the Alaska Satellite Facility before downloading the data(see instructions here).
After setting up the credentials, we can create a filesystem instance to authenticate on the remote servers in the following way (fill in the blanks):
[28]:
from getpass import getpass
nasa_fs = stac2dcache.configure_filesystem(
username=getpass("username: "),
password=getpass("password: ")
)
username: ········
password: ········
We download S1 data to the local file system:
[29]:
assets = ('browse', 'data')
for asset_key in assets:
copy_asset(
catalog,
asset_key,
filesystem_from=nasa_fs,
to_uri="./tmp"
)
[30]:
! ls ./tmp/S1B_IW_GRDH_1SDV_20190102T055723_20190102T055748_014311_01AA09_29B6-GRD_HD/
S1B_IW_GRDH_1SDV_20190102T055723_20190102T055748_014311_01AA09_29B6.jpg
S1B_IW_GRDH_1SDV_20190102T055723_20190102T055748_014311_01AA09_29B6.zip